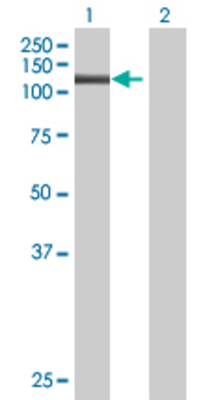
| Gene Symbol |
PMS2
|
| Entrez Gene |
5395
|
| Alt Symbol |
HNPCC4, PMS2CL, PMSL2
|
| Species |
Human
|
| Gene Type |
protein-coding
|
| Description |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae)
|
| Other Description |
DNA mismatch repair protein PMS2|H_DJ0042M02.9|PMS1 protein homolog 2|mismatch repair endonuclease PMS2
|
| Swissprots |
P54278 B2R610 Q75MR2 Q5FBX2 Q5FBX1 Q52LH6 Q5FBW9
|
| Accessions |
AAA63923 AAS00390 ABQ59090 EAW55055 P54278 AB103082 BAD89425 AB103083 BAD89426 AB103085 BAD89428 AB103086 BAD89429 AB103087 BAD89430 AI433648 AK125607 AK294661 BAG57831 AK312390 BAG35307 BC008400 BC031832 BC093921 AAH93921 BC143397 BM669686 BP251389 BQ050630 BX537558 CAD97782 CR981250 DA774159 DB527312 U14658 AAA50390 XM_006715742 XP_006715805 XM_006715744 XP_006715807 XM_011515427 XP_011513729 XM_011515428 XP_011513730 XM_011515429 XP_011513731 XM_011515430 XP_011513732 NM_000535 NP_000526 NR_003085
|
| Function |
Component of the post-replicative DNA mismatch repair system (MMR). Heterodimerizes with MLH1 to form MutL alpha. DNA repair is initiated by MutS alpha (MSH2-MSH6) or MutS beta (MSH2- MSH6) binding to a dsDNA mismatch, then MutL alpha is recruited to the heteroduplex. Assembly of the MutL-MutS-heteroduplex ternary complex in presence of RFC and PCNA is sufficient to activate endonuclease activity of PMS2. It introduces single-strand breaks near the mismatch and thus generates new entry points for the exonuclease EXO1 to degrade the strand containing the mismatch. DNA methylation would prevent cleavage and therefore assure that only the newly mutated DNA strand is going to be corrected. MutL alpha (MLH1-PMS2) interacts physically with the clamp loader subunits of DNA polymerase III, suggesting that it may play a role to recruit the DNA polymerase III to the site of the MMR. Also implicated in DNA damage signaling, a process which induces cell cycle arrest and can lead to apoptosis in ca
|
| Subcellular Location |
Nucleus.
|
| Top Pathways |
Mismatch repair, Fanconi anemia pathway
|