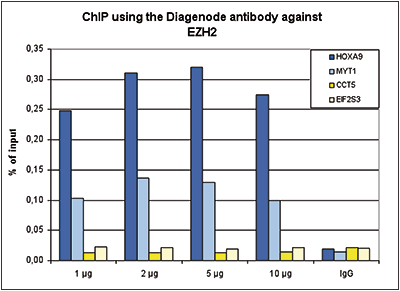
 Figure 1. ChIP results obtained with the Diagenode antibody directed against EZH2
Figure 1. ChIP results obtained with the Diagenode antibody directed against EZH2ChIP assays were performed using K562 cells, the Diagenode antibody against EZH2 (Cat. No. C15410039) and optimized PCR primer sets for qPCR. ChIP was performed with the “iDeal ChIP-seq” kit (Cat. No. C01010055), using sheared chromatin from 4 million cells. A titration of the antibody consisting of 1, 2. 5 and 10 μg per ChIP experiment was analysed. IgG (2 μg/IP) was used as negative IP control. Quantitative PCR was performed with primers for MYT1 and HOXA9, used as positive control targets, and for the coding regions of the active CCT5 and EIF2S3 genes, used as negative controls. Figure 1 shows the recovery, expressed as a % of input (the relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis).
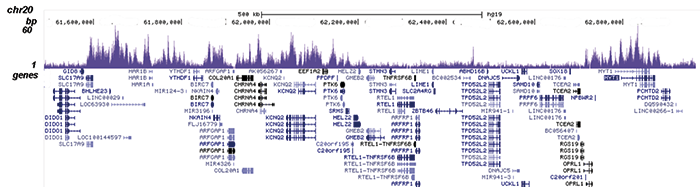
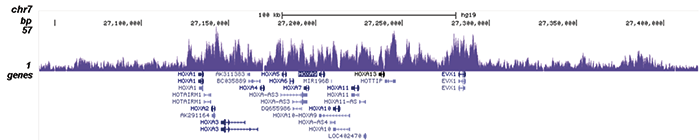
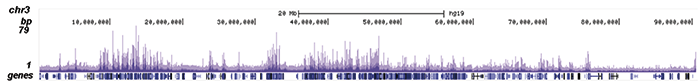 Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against EZH2
Figure 2. ChIP-seq results obtained with the Diagenode antibody directed against EZH2 ChIP was performed on sheared chromatin from 4 million K562 cells using 2 μg of the Diagenode antibody against EZH2 (Cat. No. C15410039) as described above. The IP’d DNA was subsequently analysed on an Illumina HiSeq. Library preparation, cluster generation and sequencing were performed according to the manufacturer’s instructions. The 51 bp tags were aligned to the human genome using the BWA algorithm. Figure 2 shows the peak distribution along the short arm and a 6 Mb region containing several enriched regions of human chromosome 3 (figure 2A and B, respectively), and in two genomic regions containing the MYT1 gene on chromosome 20 and the HOX cluster on chromosome 7 (figure 2C and D).
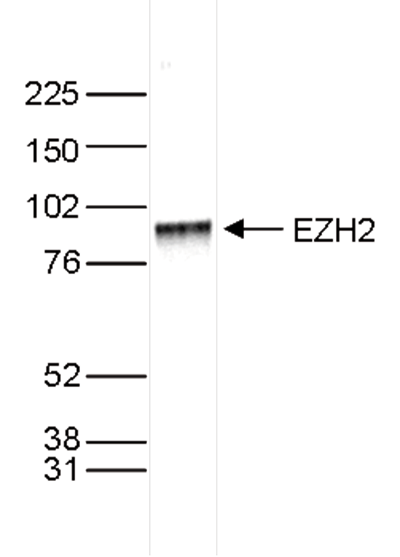
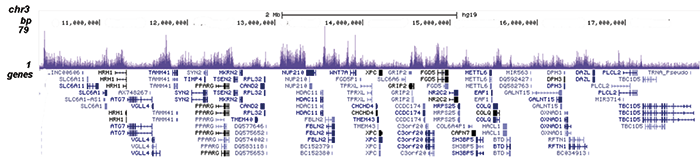 Figure 3. Western blot analysis using the Diagenode antibody directed against EZH2
Figure 3. Western blot analysis using the Diagenode antibody directed against EZH2 Nuclear extracts of HeLa cells (40 μg) were analysed by Western blot using the Diagenode antibody against EZH2 (Cat. No. pAb-039-050) diluted 1:1,000 in TBS-Tween containing 5% skimmed milk. The position of the protein of interest (expected size 85 kDa) is indicated on the right; the marker (in kDa) is shown on the left.